Overview
Small molecule drugs are our first and often last line of defense for many diseases, yet despite decades of research we still do not fully understand why a given drug works in one patient and fails in the next. These unpredictable variations are driven in part by human gut bacteria that can metabolize hundreds of drugs, altering both efficacy and side effect profiles.
To accelerate the identification of the microbial enzymes responsible for drug biotransformations, we developed SIMMER (Similarity algorithms that Identify MicrobioMe Enzymatic Reactions). This computational tool predicts gut microbial enzymes that may perform a query biotransformation (Fig. 1). SIMMER’s predictions can be used to design experiments testing candidate microbial enzymes for the proposed activity.
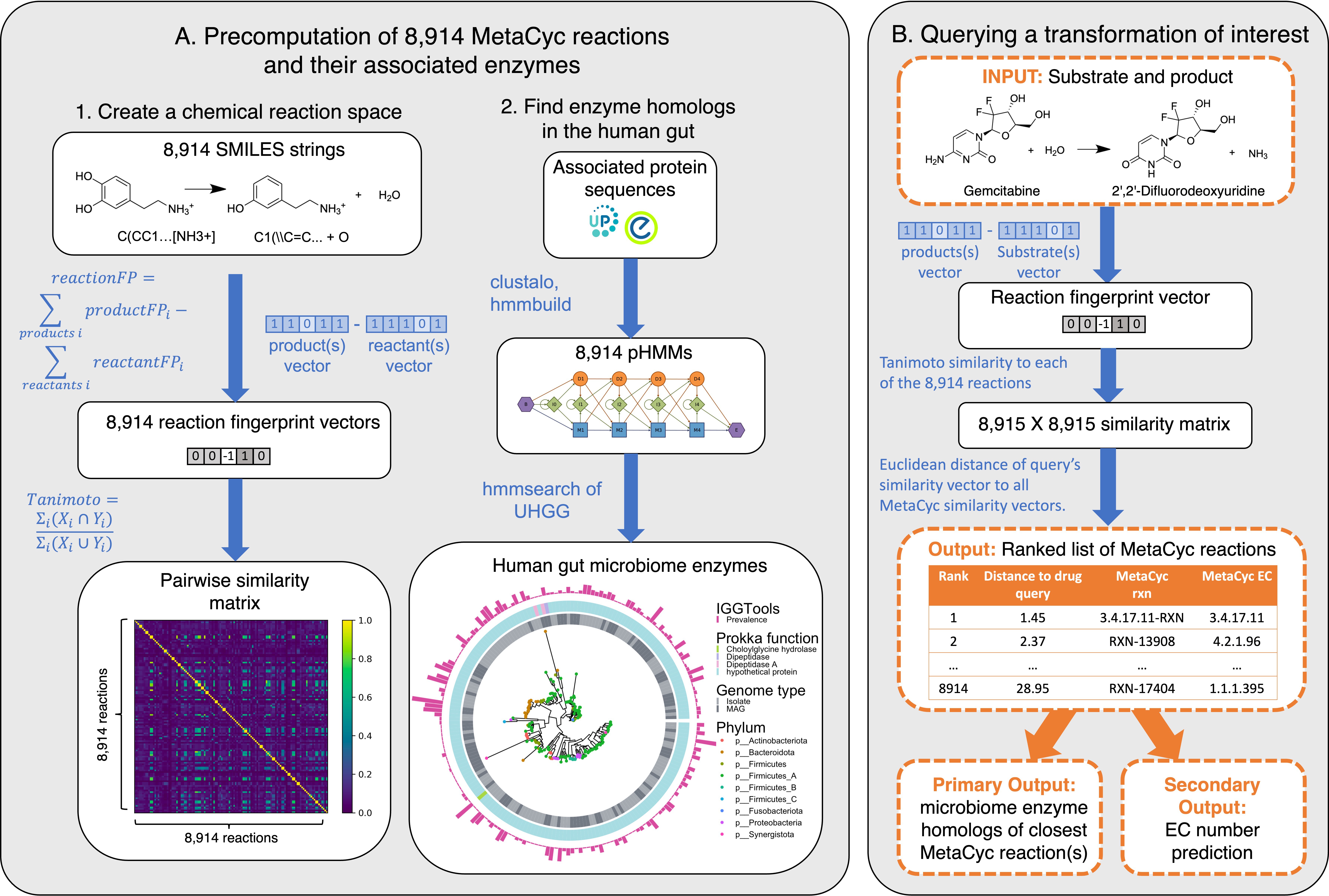
(A) Development of SIMMER: Chemical fingerprints representing 8,914 gene-annotated bacterial reactions from the MetaCyc database were created using SMILES descriptors. A latent chemical space was then computed via a pairwise reaction similarity matrix based on Tanimoto coefficients. For each reaction, relevant gene sequences were retrieved via InterPro and used to create multiple sequence alignments and subsequent profile hidden Markov models (pHMMs) using ClustalO and HMMER3, respectively. pHMMs were used to retrieve homologs in a catalog of human gut microbiome genes.
(B) Running a SIMMER query: After receiving a reaction query (input compound, cofactors, and products), SIMMER fingerprints the reaction and compares it to the precomputed chemical space from (A). MetaCyc reactions are sorted by similarity to the query. From the most similar reaction(s), human gut microbiome enzymes are predicted. Enzyme hits are reported along with their abundance and prevalence in gut microbiomes. As an auxiliary output, Enzyme Commission (EC) numbers are predicted based on enrichment in the ranked reaction list.
How SIMMER works
- Input Queries: Users input query reaction(s) as either canonical or isomeric SMILES that represent the compound, cofactors, and products.
- Reaction Matching: For each query, SIMMER identifies similar reactions from the MetaCyc database.
- Enzyme Prediction: Based on these matches, SIMMER uses the annotated enzymes to predict homologs in gut bacteria that could perform the user’s query reaction(s).
- Output: Enzyme hits with their relative abundance and prevalence in the human gut microbiome, plus a predicted Enzyme Commission (EC) number for the input reaction.
Using SIMMER
SIMMER is available for direct use on the web at https://simmer.pollard.gladstone.org/. To query more than ten input reactions at a time, or to consider more than one closest MetaCyc reaction, refer to the command-line tool documentation.
How to cite
Bustion AE, Nayak RR, Agrawal A, Turnbaugh PJ, Pollard KS. 2023. SIMMER employs similarity algorithms to accurately identify human gut microbiome species and enzymes capable of known chemical transformations. Elife 12. doi:10.7554/eLife.82401.
Software repository
SIMMER’s source code and latest updates are available on GitHub, enabling users to explore, modify, and contribute to the tool’s development. https://github.com/aebustion/SIMMER.
SIMMER web development team at Gladstone Institutes
Annamarie E Bustion, Ayushi Agrawal, Andrew Davis, Alexander R. Pico, Katherine S. Pollard
Support
For assistance, raise an issue on GitHub or contact aebustion [at] gmail [dot] com. Please note that SIMMER is actively updated, and user feedback is appreciated!